Walking the line between endosymbionts and organelles
I am broadly interested in the evolution of symbiotic organisms, organelles, and horizontal gene transfer.
In my research, I combine field work (insect and protist sampling), wet lab methods (molecular biology, cell biology, microbiology), and computational approaches (genomics, transcriptomics and phylogenomics) to understand complex multi-species symbiotic systems.
(1): Protist-bacteria and protist-archaea symbioses
Symbioses of single-celled eukaryotes are extremely poorly understood. I'm trying to shed some light on symbioses in multiple protist groups such as ciliates, diplonemids, oxymonads, and parabasalids.
(2): Symbioses of animals feeding on nutritionally poor diets (wood, plant sap, and blood)
Insects that feed on nutritionally unbalanced diets for a living often rely on symbionts to provide them with nutrients such as essential amino acids and vitamins missing from their diets. Individuals without symbiotic microorganisms (e.g. when removed by antibiotics) do not survive and reproduce. In some of these insect lineages, this up to 400 myr old symbiotic history has lead to a very intimate relationship where both the host and symbiont(s) (and their genomes) are highly adapted to the symbiosis. I am particularly interested in the origin and evolution of these symbioses and how different symbionts interact with the host, with each other, and with insect pathogens and parasites.
(2a): Protist, bacterial, and archaeal endosymbioses of 'lower' termites and wood roaches from the Cryptocercus genus
[This is the main laboratory project of my postdoctoral stay. Stay tuned as more information will appear here soon.]
(2b): Bacterial endosymbioses of scale insects (Insecta: Hemiptera: Coccoidea)
[This was my main PhD project that focused mainly on the unique tripartite nested symbiosis in mealybugs and to some level also on endosymbioses of several other scale insect families. I'm still involved in (and enjoying) several smaller projects and collaborations involving scale insects and their symbiotic microbes.]
Let me convince you that scale insects and their symbionts are cool and worth studying ;-).
There are about 8,000 species of scale insects. All of them feed on plants and some of them are notorious cosmopolitan pests. Almost all of the 8,000 species have very diverse and poorly understood consortia of bacterial or fungal symbionts.
Perhaps most fascinating scale insects are mealybugs (Pseudococcidae) because they are the only group of animals with a stable tripartite nested bacterium-within-bacterium-within-insect-cell symbiotic system. Apart from having the unique symbiotic system, they also have absolutely crazy genetic systems. They are so called obligate chimaeras, their body develops not from a single fertilized egg like bodies of most animals, but from two genetically different cells. One of these cells develops into a specialized organ that houses symbiotic bacteria. That's not all. They are also very mean to genetic information encoded on chromosomes from males. The phenomenon is called pseudo-arrhenotoky or paternal genome elimination (PGE) and means that paternal chromosomes are silenced in somatic tissue and eliminated from the germ line and thus not passed on their offspring.
Still not convinced that mealybugs are awesome? Read my two papers showing that horizontal gene transfer from several different bacteria contributes to functioning of the tripartite-nested symbiosis and that the innermost symbionts were replaced several times over evolutionary history.


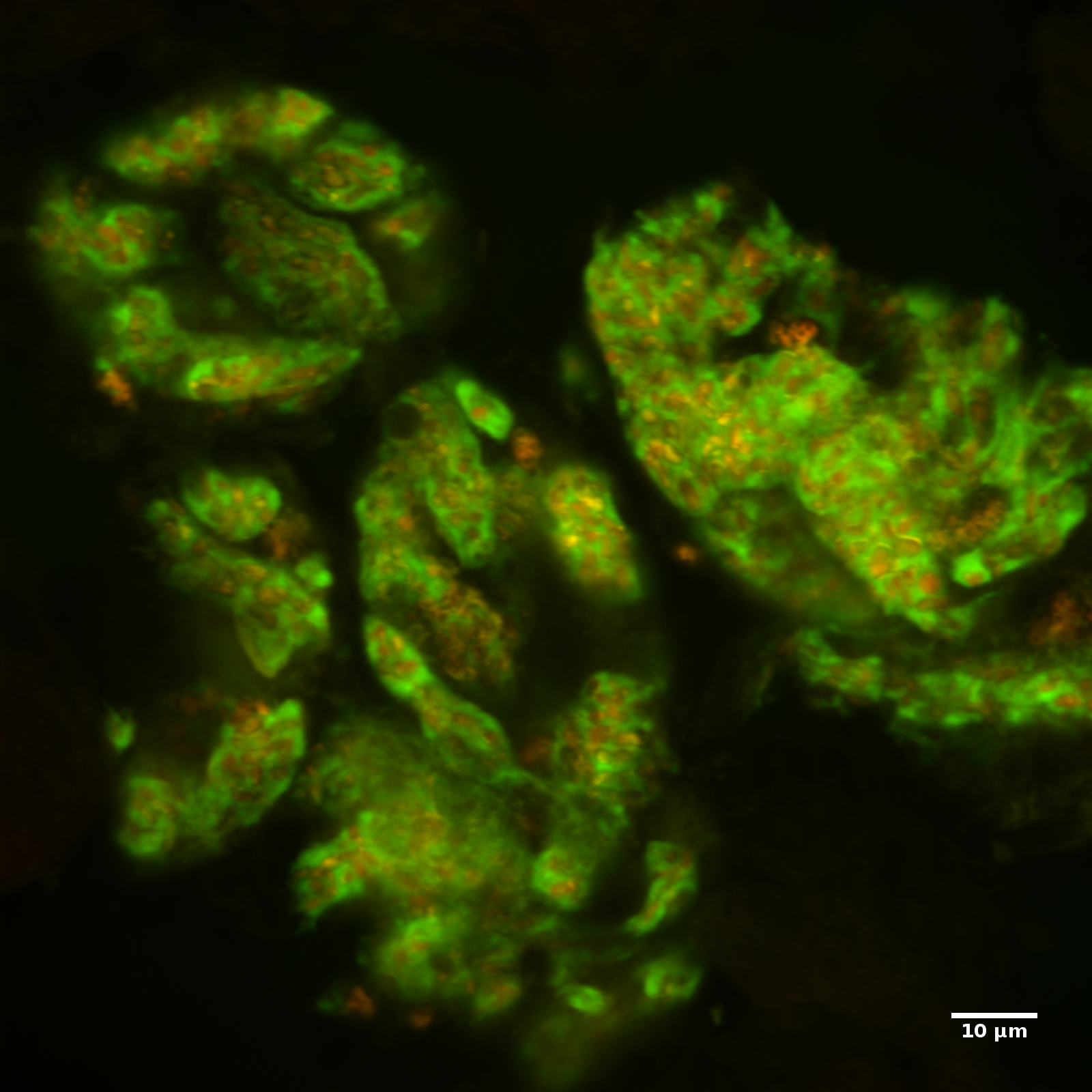
Left: Citrus mealybug (Planococcus citri): a winged male and a nymph at crawler stage. Center: A mature female and a second instar nymph [image by Alexander Wild]. Right: Fluorescence in situ hybridization of citrus mealybug bacteriocytes with Moranella endobia cells (in orange) inside Tremblaya princeps (in green) cells.
(2c): Bacterial endosymbioses of lice, louse flies, bat flies, and other bloodsucking insects
[I'm no longer involved in any projects focused on symbioses of blood-feeding animals. These symbioses are more or less only a (strange) hobby for me now, so see my publications or contact me directly if interested in collaborations etc.]